The Short Version
Brewing practices in both home and commercial breweries have undergone somewhat of a revolution over the past decade, leading to a cohort of brewers who approach brewing from a much more technical & microbiological perspective. As a direct consequence of this, some commercial yeast products have been revealed to be other than what the manufacturers have stated - in at least some cases, with the manufacturer themselves being unaware that their product was a yeast/bacteria different from what they believed they had. In this blog post we reveal that the yeast sold by White Labs as Brettanomyces vrai (WLP648) - ironically a yeast mis-identified previously by the same manufacturer - is, in fact, a blend of two different yeasts - both are Brettanomyces bruxellensis, but are separate strains...although strains which appear to have evolved from a recent common ancestor.Some Background
Brewing practices have changed dramatically over the past decade, with procedures such as sour worting, wild captures, and home/brewery isolated yeasts going from rare experiments to commonplace brewing practices. This change in brewing practices has led to some issues with commercially sourced yeasts - as one example, the growth of practices such as sour worting have revealed yeast-contamination issues in packaged "pure" strains of Lactobacillus. Similarly, the more microbiology-centric practices of home and commercial brewers has led to some unexpected revelations, including identification of "Brettanomyces trois" as a unusually flavourful strain of conventional brewers yeast (Saccharomyces cerevisiae). I was part of that effort, and the results of my and others work in identifying this yeast are the subject of a previous post. According to the manufacturer, this mis-identification was due to a chance contamination of "Brett trois" by this strain of Sacc, leading to the release of the "correct" strain of Brettanomyces, under the 'vrai' (French for 'true') strain name.But is the strain name accurate - is this truly a pure strain of Brettanomyces? Most of us assumed so, even though this strain shows some characteristics when used as a pure culture for primary fermentation that run contrary to how most Brettanomyces behave when used for primary fermentation. When used in primary fermentation, most Brettanomyces act much like Saccharomyces - they rapidly ferment the wort, usually leave some residual sugars behind, and don't evolve over ageing as much as beers do when Brettanomyces are added during secondary fermentation - e.g. there is a lack of phenol production and super-attenuation. Beers brewed with WLP648 do ferment out fairly quickly, but tend to be more highly attenuated than beers brewed with other strains of Brettanomyces as the primary yeast. In addition, beers brewed with WLP648 also show some development during ageing similar to that of beers with Brettanomyces added to secondary - i.e. emergence of phenolic "funk", and additional attenuation of the beer. So is WLP648 simply a more aggressive Brettanomyces than other common brewing strains, or is something else going on?
To our knowledge, it was assumed by other brewers that Brett vrai was simply a somewhat more attenuative strain of Brett - that is - until my friend and brewing collaborator (and co-author of this blog post) Devin streaked WLP648 on a wort-agar plate. Initially, the plate appeared as one would expect of a pure culture - all colonies on the plate appearing similar in size, shape and colouration. But over a longer incubation time smaller colonies began to appear between the larger colonies, leading us to speculate that there may be a second strain of yeast in WLP648.
Using a combination of classical microbiology, microscopy, gene sequencing and test batches, Devin and I explored the two strains of yeast present in WLP648, demonstrating that Brett vrai contains two unique strains of Brettanomyces bruxellensis, strains which share a relatively recent common ancestor, but are otherwise quite different in their morphology and brewing characteristics.
Experimental details can be found below the fold.
First Signs of Contamination
 |
| Figure 1: WLP648 on malt-agar |
 |
Figure 2: Micrograph of WLP648
|
 |
Figure 3: Malachite-green stained pure cultures of LCV
(left) and SCV (right).
|
Although it is hard to believe that the high proportion of SCV's in Devin's tube of yeast could have come from contamination when he opened the tube, or was a one-off contamination during packaging by the manufacturer, or due to mis-handling by the home brew shop where the yeast was purchased, it was necessary to confirm the presence of both the LCV and SCV in a second lot of WLP648. As such, we purchased a second tube of WLP648 from a different homebrew shop, confirming with the shop owner that the yeast was of a different lot than the first lot used (Table 1). This tube was cleaned externally with Wescodyne (a lab-grade germicidal agent), opened in a sterile culture hood, and plated on freshly autoclaved media. Both the LCV and SCV were present in this second lot of WLP648, present in the manufacturers' tube at roughly the same proportions, showing similar growth kinetics, and with similar morphology in micrographs (not shown).
Table 1: WLP648 lot numbers and expiry dates
| Brewer | Yeast Lot Number | Expiry Date |
| Devin | 1030320 | May 22, 2017 |
| Bryan | 1029514 | April 30, 2017 |
Species Identification by ITS Sequencing
 |
Figure 4: ITS PCR of LCV
(middle) and SCV (right).
Left lane is a size-
standard.
|
Next, a firm genetic identification of LCV and SCV was performed using ITS ribosomal sequencing, as outlined in my previous blog posts (1, 2, 3). The first step of this process is to use PCR to amplify a specific portion of the yeast ribosome termed the "Internal Transcribed Spacer" (ITS) region. This is the preferred DNA region for identifying yeast species, as it tends to evolve at a rate where it varies minimally within a species, but varies sufficiently between species to allow for species identification. The ITS of both the LCV and SCV amplified well, producing a DNA fragment of the expected ~800 base-pair size (Figure 4).
The resulting DNA molecules were then purified and submitted for sequencing, and the resulting sequences BLASTed to search for the closest match in the NCBI database, as well as aligned against my personal collection of representative ITS sequences (Table 2, Figure 5).
| Strain | ITS Sequence | NCBI ID |
| LCV | CCGTAGGTGA ACCTGCGGAA GGATCATTAC AGGATGCTGG GCGCAAGCCC GTGCAGACAC GTGGATAAGC AAGGATAAAA ATACATTAAA TTTATTTagt tTagtCAAGA AAGAATTTTA AAACTTTCAA CAATGGATCT CTTGGTTCTC GCGTCGATGA AGAGCGCAGC GAATTGCGAT ACTTAATGTG AATTGCAGAT TTTCGTGAAT CATCGAGTTC TTGAACGCAC ATTGCGCCCT CTGGTATTCC GGAGGGCATG CCTGTTTGAG CGTCATTTCC TTCTCACTAT TTAGTGGTTA TGAGATTACA CGAGGGTGTT TTCTTCAAAG GAAAGAGGGG AGAGTGAGGG GATAATGATT TAAGGTTTCG GCCGTTCATT ATTTTTTTCT TCTCCCCCAG TTATCAAGTT TGACCTCAAA TCAGGTAGGA GGACCCGCTG AACTTAAGCA TATCAATAAG CGGA | Brettanomyces bruxellensis (100% identity) |
| SCV | TCCGTAGGTG AACCTGCGGA AGGATCATTA CAGGATGCTG GGCGCAAGCC CGTGCAGACA CGTGGATAAG CAAGGATAAA AATACATTAA ATTTATTTAG TTTAGTCAAG AAAGAATTTT AAAACTTTCA ACAATGGATC TCTTGGTTCT CGCGTCGATG AAGAGCGCAG CGAATTGCGA TACTTAATGT GAATTGCAGA TTTTCGTGAA TCATCGAGTT CTTGAACGCA CATTGCGCCC TCTGGTATTC CGGAGGGCAT GCCTGTTTGA GCGTCATTTC CTTCTCACTA TTTAGTGGTT ATGAGATTAC ACGAGGGTGT TTTCTTCAAA GGAAAGAGGG GAGAGTGAGG GGATAATGAT TTAAGGTTTC GGCCGTTCAT TATTTTTTTC TTCTCCCCCA GTTATCAAGT TTGACCTCAA ATCAGGTAGG AGGACCCGCT GAACTTAAGC ATATCAATAA GCGGAGGAAA GGATCATTAC AGGATGCTGG G | Brettanomyces bruxellensis (100% identity) |
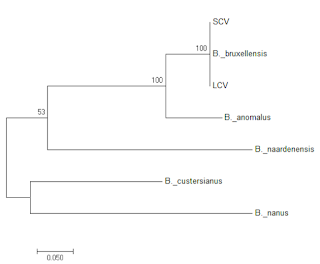 |
Figure 5: Phylogenic tree of LCV and SCV relative to representative
ITS sequences from a range of Brettanomyces species.
|
So both strains are clearly and unambiguously Brettanomyces bruxellensis, leading to the question of whether we are looking at petite mutants, or whether they are different strains.
Strain Identification by RAPD PCR
 |
Figure 6: RAPD PCR of LCV and
SCV with the OPK3, OPD19 and OPC20 primers. Left lane is a size- standard. |
The procedure is fairly simple; a each PCR reaction is run with extracted DNA and one of the above primers. The 35 PCR cycles are run using a 94C denaturation step (1 min), 35C annealing step (1 min) and a 2 min elongation at 72C. For those not familiar with PCR, that means we are "photocopying" regions of the genomic DNA, using a low annealing temperature to pick up differences between strains (higher annealing temperatures copy specific regions of DNA, lower is less specific).
 |
| Figure 7: Intensity plots of RAPD PCR gel in Figure 6. |
What does this mean? The near-identical OPK3 and OPD19 banding pattern means that these strains are close-cousins, rather than radically different strains. Indeed, this shared banding pattern is consistent with a recent common ancestor in the past century or so. But the presence of unique bands in the OPC20 lanes tells us that these two strains of yeast have differentiated from each other fairly significantly.
In other words, these are unique strains of Brettanomyces, and not merely minor variations (i.e. petite versus non-petite) within a single strain. This also eliminates the possibility that this is an environmental contaminant Devin or I picked up when opening the tubes - the likelihood of having such a similar strain of Brettanomyces floating around both of our personal breweries (and my workplace) is negligable, meaning the only place that these two yeasts could have come from is the white-labs vial.
Tasting Notes
 |
| A rare stove-top brew |
Tasting took place on May 9th, with both Devin and I sampling the beers side-by-side:
 |
| Left: SCV, Right: LCV |
- FG: 1.014, 5.3% ABV
- Attenuation: 73%
Aroma: The aroma of this beer was amazing - an intense dark cherry aroma dominated, with a touch of barnyard or wet hay-like funk in the background.
Flavour: The flavour of this beer matched the aroma; intense fruit character with strong notes of dark cherry and red wine. Some funk flavours were present in the background. Finish was sweet and refreshing.
Mouthfeel: Medium-bodied and whetting. After taste was sweet and fruit, with a noticeable acidity.
Overall: For such a simple beer, this one turned out great. Fruity with a hint of funk; I could see using this yeast in a darker farm house ale, or perhaps in some sort of a kettle sour.
LCV:
- FG: 1.016, 4.9% ABV
- Attenuation: 69%
Appearance: Identical to the SCV, an off-putting copper-grey with minimal head.
Aroma: The aroma of this beer was quite different; an intense mousy/musty aroma dominated, with only the faint aroma of fruit in the background.
Flavour: Here Devin and I disagree. I found the flavour to be an overwhelming, to the point of being unpleasant, mousy/stale funk, behind which was a mild "generic fruit" character. Devin was less negative about the character, although we both agreed on the particular flavours present. Finish was sweet, but with a strong funk note.
Mouthfeel: Much the same as the SCV; medium-bodied and somewhat refreshing.
Aroma: The aroma of this beer was quite different; an intense mousy/musty aroma dominated, with only the faint aroma of fruit in the background.
Flavour: Here Devin and I disagree. I found the flavour to be an overwhelming, to the point of being unpleasant, mousy/stale funk, behind which was a mild "generic fruit" character. Devin was less negative about the character, although we both agreed on the particular flavours present. Finish was sweet, but with a strong funk note.
Mouthfeel: Much the same as the SCV; medium-bodied and somewhat refreshing.
Overall: To my palate, the funk of the LCV strain was overwhelming and unpleasant; I would not drink this beer, and if served it in a pub would send the pint back. Devin was less negative, but also thought that the LCV was a poor cousin to the SCV.
Blended Strain: I wasn't smart enough to ferment the test beer with both strains, but luckily Devin had a bottle of a similar beer made with Vrai (the same tube of Vrai the SCV and LCV were isolated from). I didn't take complete notes on Devins beer, but it very much split the middle between the LCV and SCV strains - mid-road on both the cherry and funk notes. One aspect that was unique was a skunky character that Devin noticed built over time. This was not the skunky character of light-struck hops, but rather a bona fide intense aroma much like that of musk. Again, I preferred the SCV over the blend, but the blend made a good beer.
I also brewed an IPA with Vria...which will be the topic of a future post. TLDR version would be that Vrai ruined my IPA.
Conclusions
What are They?Given the growth and genetic differences, and the higher attenuation of the SCV compared to the LCV, we can conclusively state that these are not normal versus petite mutants of a single strain. Rather, these are two unique, but closely related strains. While they are minimally different genetically, the genetic changes they have undergone have led to a number of large changes in biology. Assuming the SCV descended derived from the LCV - the most likely interpretation of the RAPD data - these mutations led to a large decrease in cell volume, a change in the cell division or adhesion properties of the yeast, and led to lower POF gene activity (funk), higher expression of one or more of the 6 or so genes associated with ester production Atf1p, Atf2p, Eht1p, Eeb1p, EEB1 and Iah1p), and influenced some of the two dozen or so genes which affect cell volume.
Without a full genome sequence it is not possible to state exactly what occurred, but the fact that the SCV varies genetically from the LCV by the addition of between two to four OPC20 sites, it is most probable that the SCV is a descendant of the LCV, with the differentiation between the strains a product of the insertion of genetic materials. Again, without sequencing this is supposition, but insertions of new DNA is commonplace in yeast, usually occurring via transposons. Tansposons are mobile genetic elements which can made copies of themselves elsewhere in the genome. This ability to "copy" themselves means that they are common in the genome - your genome is ~44% transposon - but it also means that they can greatly influence the regulation of genes. In yeast, movement of transposons is fairly common, and is associated with large-scale changes in gene expression, and at least in the case of Saccharomyces, transposon-associated changes in gene expression account for some of the differences between yeast strains.
How Did White Labs Miss This?
I don't know what White Labs does behind the scenes to check their strains, but the fact that this has missed their attention (assuming the blend is not deliberate) is not surprising to me. Firstly, it was extremely difficult to separate the SCV from the LCV as the larger number of SCVs, combined with their slower growth rate, meant that each LCV colony grew on top of several SCV colonies. As such we had to go to great lengths to get a pure culture. Secondly, the much slower growth rate of the SCV meant that unless you specifically went out of your way to culture the plate for an extended period of time, you would not see the SCV colonies; even though they were mixed in with the LCVs.
Does it Matter?
From a brewery perspective, this probably doesn't matter. While the SCV produces a nicer beer (IMO; I love the intense cherry flavour), the blend itself produces a beer with a more balanced fruit:funk character. From a brewing-management position, the slower growth-rate of the SCV means that the characteristics of beers brewed with re-pitches of this blend may vary depending on when you repitch - yeast taken at high kraussen or soon after kraussen has fallen will be LCV-dominated, while beer that has aged for a while will have a more equal, or even SCV-dominated population. This may impact the overall flavour profile of the beer, although further experimentation will be required to see if that is the case.
Awesome post.
ReplyDeleteReminded me of this post. https://brettanomyces.wordpress.com/2009/04/06/two-for-one-brettanomyces-strains/
DeleteI still have to test if whitelabs lacto cultures have yeast in them, several posts outline that it might but maybe it could be contamination. It'd be cool if you could do the same with WLP645. I'm pretty sure chad noted years back that it had pichia in it (he posted on your blog during the wlp644 saga :)).
DeleteAFAIK, Drai and Vrai are the same strain, so this "contamination" likely goes back to the origonal source of the yeast. Personally, I'll only be using the SCV moving forward.
DeleteIn terms of contamination of lacto cultures, we know that is an issue with WL, with several people finding that. My 645 stock appears to be clean, but I also cleaned it up from a mixed culture I was given, so that may not be representative of the commercial product.
This comment has been removed by the author.
ReplyDeleteNice work gentlemen. I will be isolating the SVC for attempted Brett IPA.
ReplyDeleteI wish I had the skills to isolate and culture my own SCV. I like 648, it's one of my favorite Brett pitches, but reading this makes me realize what I like can be attributed to SCV.
ReplyDeleteI'll get there one day, but at the moment I am not well suited to sterile operations like culture plates and isolates.
I ordered my vial of WLP648 in 2015, lot #1016078. Under the scope, I did not notice the two morphological variants that you did, but did find lacto in there as well. In the email that I sent to Sarah Neel at white labs, I said "I initially streaked this culture onto malt plates, but had issues with growth of rod shaped bacteria. Additionally, I observed motile rods in the culture received under 1000X magnification that resembled the bacterial colonies that grew up on malt agar.
ReplyDeleteI subsequently streaked the culture onto WLN, WLD, and MRS plates. I was able to isolate yeast colonies, but still saw a significant number of lactobacillus colonies on MRS. (Along with petite colonies that I did not do wet mounts for) While I was able to isolate what I believe is the desired Brettanomyces in the end, I am writing to inquire if any other brewers have had issues with contamination in this lot? Knowing the source of contamination either on my end or yours would be helpful for future isolations."
Her reply was that there was no contamination on their end. I don't have any big point to make, except that they had at least heard that there is more than one thing in 648.
I've not heard of any contamination issues in this yeast (although yeast contamination of their lacto & pedio cultures is not unheard of).
DeleteI'm not sure what bacteria you saw, but if it was motile it was not lactobacillus. Devin and I have been in contact with White Labs; they think that the two yeast colony types may be due to genetic instability in the strain, although the fact that we have pure strains of SCV and LCV that propagate without formation of the other strain type suggests against that.